Discovering Drug-related Adverse Events from Social Media Using Topic Models
Problem: early detection of drug-related adverse events
According to the Food and Drug Administration (FDA), an Adverse Event (AE) is "any undesirable experience associated with the use of a medical product." Early detection of drug-related adverse events benefits not only the drug regulators and manufacturers for pharmacovigilance, but also patients as certain adverse events can be life-threatening.
In order to capture AEs and Adverse Drug Reactions (ADRs), various surveillance systems—spontaneous reporting systems (SRSs)—have been developed around the world. In the US, The FDA’s Adverse Event Reporting System (AERS) is a major SRS with more than four million reports. In Europe, the European Medicines Agency developed the EudraVigilince. The World Health Organization has an international pharmacovigilance system as well. Aforementioned SRSs (particularly AERS) contain many reported post-market drugs' AEs. Although drug manufactures are required to report all known AEs, the majority of adverse events are detected by physicians and patients, where reporting is voluntary. Thus, the overall incidences of AEs can be significantly underestimated.
Method: discovering AEs through mining social media
Social media especially Twitter, given its deep roots in people's daily lives, has a great potential of becoming a new data feed for real-time pharmacovigilance. For example, from such a Twitter message (i.e., tweet) "this warm weather + tamoxifen hot flushes is a nightmare!" we can infer the possible drug use ("tamoxifen") and associated side effects ("hot flushes") about the tweet publisher. The knowledge discovered through social media mining can certainly be used to identify new adverse events in a timely manner and to confirm those adverse events reported through existing mechanisms.
In [1], we proposed an analytic framework for detection of drug-related AEs using tweets. Specifically, we built Support Vector Machine (SVM) models with textural and ontological features extracted from tweets. However, the feature extraction methods we applied are not designed for tweet-like social media contents that often contain 1) misspellings, odd abbreviations and non-word terms, and 2) fragmented and ungrammatical sentences. Thus, the identification accuracy of the models is not satisfying ( 74%). In this work, we employ a powerful topic modeling method for feature extraction.
Technique I: topic model based feature extraction
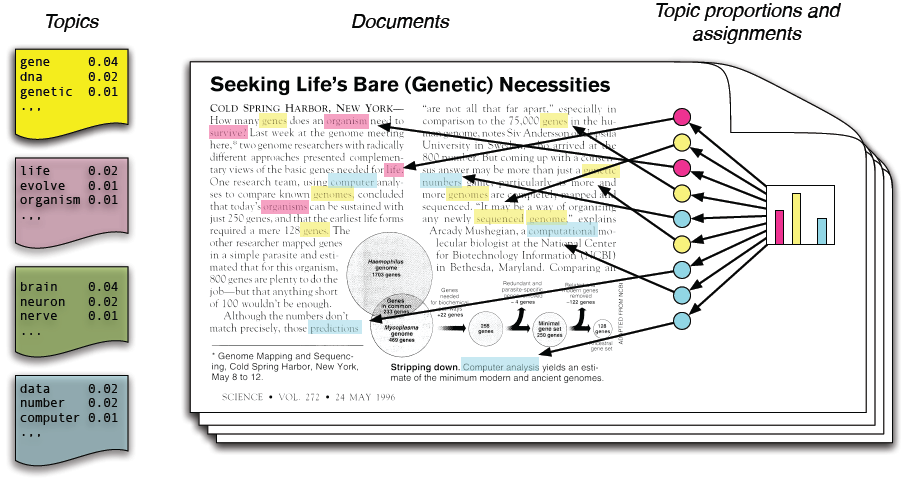 Figure 1: Illustration of the intuitions behind latent Dirichlet allocation (Courtesy of CACM)
Figure 1: Illustration of the intuitions behind latent Dirichlet allocation (Courtesy of CACM)Topic models are statistical algorithms that can discover the main themes from a large unstructured collection of documents by analyzing the words of the texts. Topic models can organize the collection according to the discovered themes. One of the most important features of topic models is that they do not require any prior annotations or labeling of the documents—the topics emerge from the analysis of the original texts. A very nice introduction to topic modeling by David Blei was recently published at CACM.
Latent Dirichlet Allocation (LDA) is the simplest topic model. The intuition behind LDA is that documents exhibit multiple topics. For example, consider the article in Figure 1. The article "Seeking Life's Bare (Genetic) Necessities" is about using data analysis to determine the number of genes an organism needs to survive. The article comprises multiple topics such as data analysis (words highlighted in blue: data, number, computer) and genetics (words highlighted in yellow: dna, gene, genetic). LDA can be used to discover those topics in an unsupervised manner.
Previous study has shown that LDA based feature selection is reliable and generally better than document frequency based feature selection. In this study, we apply the LDA model to categorize the collection of tweets into latent topics. In the LDA model, each document (tweet(s) in our study)—treated as a vector of word counts using the bag-of-words approach—is viewed as a mixture of probabilities over the topics, where each topic is represented as a probability distribution over a set of words. We then use the probability distribution of topics as features to train the SVM prediction models.
Before applying the LDA algorithm, we first sanitize the dataset by removing the hyperlinks, hash-tags "#" and reply-tags "@", and then apply the WordNet Lemmatizer in the NTLK to group the different inflected forms of a word into its lemma. By doing so, we create a "clean" dictionary for the LDA model.
Technique II: AEs discovery through mining tweets
The process of discovering AEs from tweets has two parts: 1) identifying the users of the drug of interest, and 2) finding possible side effects attributed to the use of the drug. In both parts, an SVM classification model is built based on features extracted from tweets. The overall process of tweets mining and performance improvement by using LDA based feature extraction can be found in the poster below.
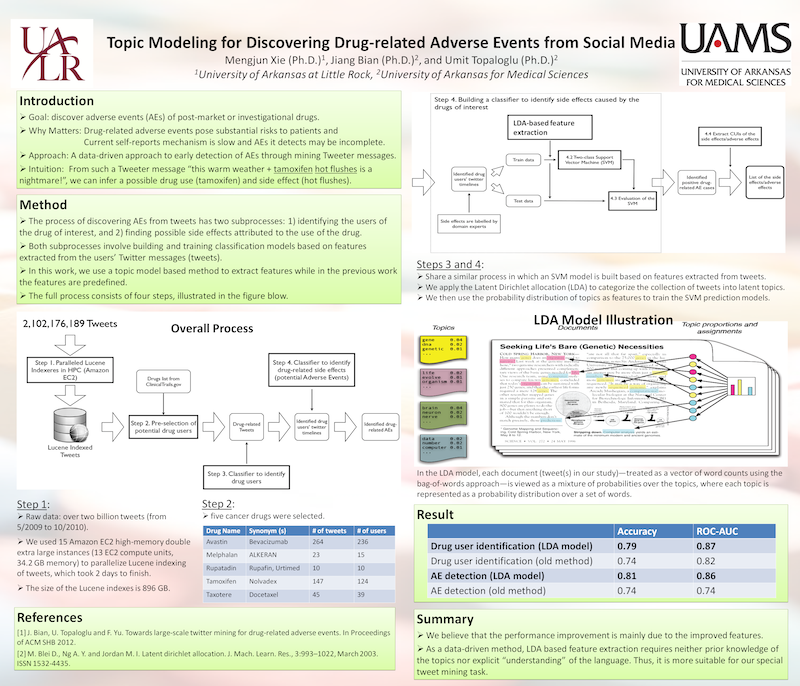 Figure 2: The poster presented at AMIA TBI 2013 (Full Size)
Figure 2: The poster presented at AMIA TBI 2013 (Full Size)Results
The performance of both SVM models has been much improved compared to the previous study. The mean accuracy of the model for identifying drug users is 79% (ROC-AUC value is 0:87). For the AE classification model, the accuracy is 81% (ROC-AUC is 0:86). We believe that the performance improvement is mainly due to the improved features.
In the previous study, the ontological features are extracted using MetaMap based on UMLS Metathesaurus concepts. The MetaMap’s NLP models are trained on biomedical literature that 1) contain less language errors and 2) are formulated with standard medical terminologies. However, Twitter users have a very different vocabulary and their writing styles are usually informal. In this study, we apply the LDA model directly to the tweets without introducing a different vocabulary. Furthermore, the methods for obtaining textual features in the previous study require manual analyses of the tweets to identify language features related to the tasks. In contrast, LDA is a data-driven method, where neither prior knowledge of the topics and nor explicit "understanding" of the language is required.
Project Members
- Mengjun Xie, Computer Science @UALR
- Umit Topaloglu, Biomedical Informatics @UAMS
- Yu Fan, Research Program, Information Techonology @UAMS
- Jiang Bian, Biomedical Informatics @UAMS
Tools used for this study
- Gensim - Topic Modelling for Humans. Gensim is a Python based software suite for topic modeling. A very nice tutorial of using gensim for topic modeling (including LDA) is also provided there.
- MetaMap
Source Code
The source code can be accessed at github.com/mengjunxie/ae-lda under the MIT license.